SNP Arrays for Cytogenetic Analysis
 A SNP (Single Nucleotide Polymorphism) is a position in the genome where the base-pairs that compose DNA differ between people.
A SNP (Single Nucleotide Polymorphism) is a position in the genome where the base-pairs that compose DNA differ between people.
- SNP arrays use known DNA sequences in regions of known SNPs as the probes.
- SNP arrays can detect mutations/polymorphisms in a gene sequence that may be constitutional (patient specific) or somatically acquired (disease specific)
- SNP arrays can also be used to analyse copy number & Loss of Heterozygosity (LOH) – including copy-neutral Uni-parental Disomy (UPD)
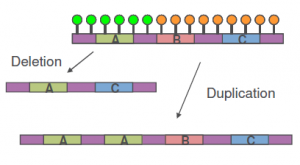
- CNVs (Copy Number Variations) are discrete regions of the genome where the copy number deviates from the normal (n=2)
- CNVs are identified using intensity information from consecutive SNPs/probes
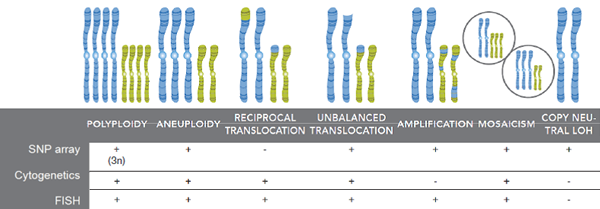
- SNP arrays can be used as an alternative to cytogenetic screening, with additional detection of LOH and UPD, however arrays cannot detect balanced translocations therefore other methods (PCR / FISH) must also be used (see summary below)
Illumina’s Infinium chemistry is used for SNP array sample processing
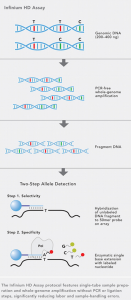
Infinium chemistry is based on single base extension of whole genome amplified genomic DNA, followed by fragmentation.
Infinium chemistry utilises the single probe sequence (50mer) designed to hybridise immediately adjacent to the loci of interest, stopping one base before the interrogated marker.
Enzymatic single-base extension following hybridisation incorporates a labelled nucleotide to ensure marker specificity.
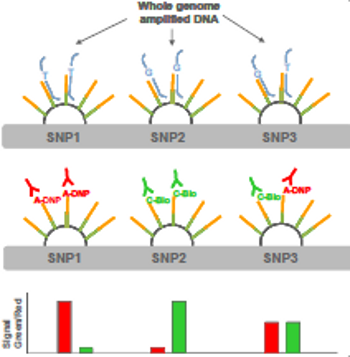
Homozygotes are indicated by red/red (eg SNP1) or green/green signals (eg SNP2), and heterozyotes are indicated by combined red/green signals (eg SNP3) visualised as yellow on capture.
The figure below shows the diagrammatic representation of the allele ratio and the image to the right shows the scanned image.
Subsequent dual-color florescent staining allows the labeled nucleotide to be detected by Illumina’s BAR or iScan imagining system, which identifies colour and signal intensity. For genotyping assays, the red and green colour signals specify each allele.
SNPs are Dual Purpose Markers:
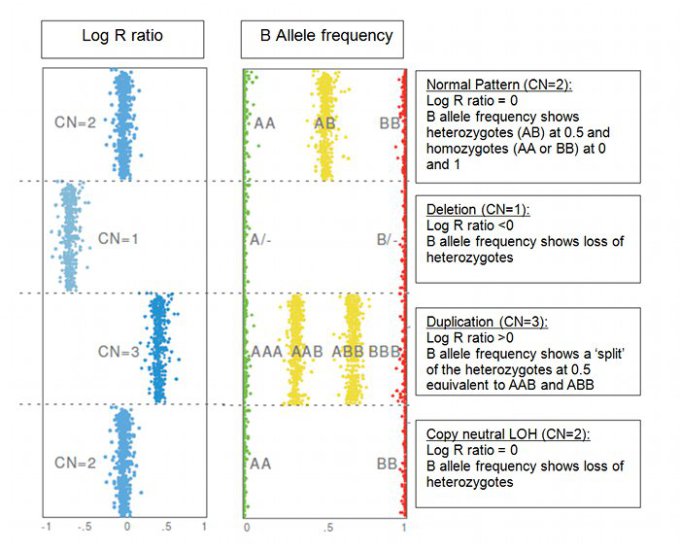
- The log R ratio shows the intensity signals of the SNPs, which indicates gains or losses relative to normal (0)
- B allele frequency gives information about the genotype as a measure of allelic ratio
- The information is combined to provide copy number (CN) information; examples of which are shown in the figure below