 Fluorescent in site hybridisation (FISH) is a method for detection structural abnormalities of chromosomes. These include translocations, where breaks have occurred at the same time in two chromosome and the wrong ends have joined, and deletions and amplifications, where parts of a chromosome have been lost from the cell or multiple abnormal copies have been produced.
Fluorescent in site hybridisation (FISH) is a method for detection structural abnormalities of chromosomes. These include translocations, where breaks have occurred at the same time in two chromosome and the wrong ends have joined, and deletions and amplifications, where parts of a chromosome have been lost from the cell or multiple abnormal copies have been produced.
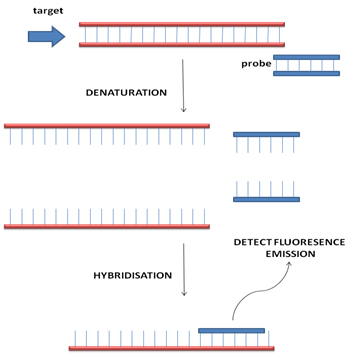
FISH uses probes based on DNA sequences labelled with fluorescent dyes. The DNA sequences are chosen to hybridise specifically to localised area of a particular chromosome of interest. The presence of the hybridised probe in the cell nucleus can be visualised using a fluorescent microscope. This is a versatile technique that can be applied to almost all type of specimen and most importantly sections from fixed tissue biopsies.
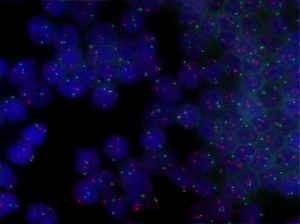
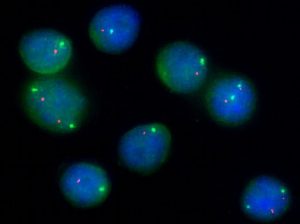
Usually multiple probes labelled with different fluorescent dyes are used in parallel and comparison of the distribution of the signals from each probe is the basis of diagnosis. In a normal cell two signals, one for each of the pair of chromosomes being studied, will be seen. Co-localisation of signals from probes that normal hybridise to different chromosomes would be seen if a translocation is present. Absence of a signal would suggest that a segment of chromosome has been deleted.
FISH is a currently used in a number of critical application including the diagnosis of Burkitt Lymphoma and identification of deletions of TP53 in chronic leukaemia.
Examples