Bead Array Technology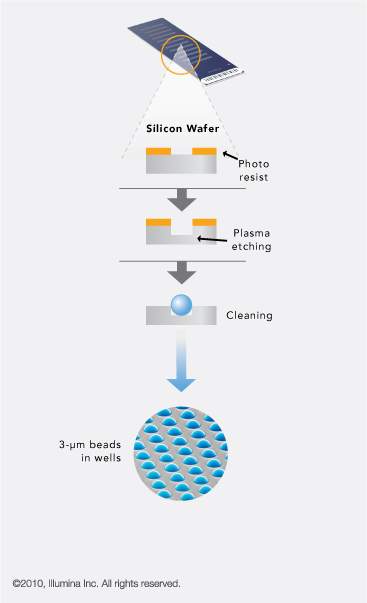
Microarray technology allows for whole genome analysis of patient samples using a CHIP-based format in the investigation of gene expression and genotyping/copy number variation (CNV) analysis, using a multiplex approach.
Depending on the type of array for investigation, genomic DNA or total RNA extracted from the tissue samples are used as the starting material that will ultimately be hybridised to the probes on the array. Fluorescent markers are incorporated into the hybridised sequence during the protocol (one or 2 fluorochromes depending on the downstream application). The fluorescence is then imaged and quantified during scanning.
Illumina use a bead array format, in which multiple copies of each probe are attached to a bead type. The beads are randomly aligned onto the array, and are identified during imaging using a specific address code for each probe/bead type and unique decode files specific for each individual array.
 Illumina BeadArray Technology uses 3-micron silica beads that randomly self assemble in microwells etched
Illumina BeadArray Technology uses 3-micron silica beads that randomly self assemble in microwells etched
on planar silica slides.
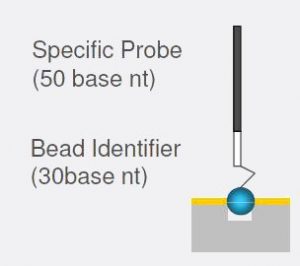
 Each bead is covered with hundreds of thousands of copies of the same specific oligonucleotide probe that act as the capture sequences in one of Illumina’s assays. These multiple copies, combined with multiple beads of each type per array, procides a high degree of redundancy. The bead identifier is used as an address label to decode the beads locations on the chip.
Each bead is covered with hundreds of thousands of copies of the same specific oligonucleotide probe that act as the capture sequences in one of Illumina’s assays. These multiple copies, combined with multiple beads of each type per array, procides a high degree of redundancy. The bead identifier is used as an address label to decode the beads locations on the chip.
Illumina BeadArray technology is utilized for a broad range of DNA and RNA analysis applications, including gene expression (DASL) and genotyping/copy number variation (CNV) analysis (SNP arrays), currently in use in HMDS.
Gene Expression Arrays
- Investigate level of expression (mRNA) of genes of interest or from the whole genome
- Use cDNA probes on the array
- Utilise Hierarchical clustering to define sub-categories within a cohort of samples, eg. Diagnostic / prognostic sub-groups
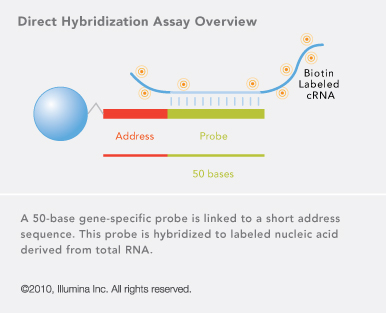
Direct Hybridization for intact RNA
- Illumina direct hybridisation protocols feature a first- and second-strand reverse transcription step, followed by a single in vitro transcription (IVT) amplification that incorporates biotin-labelled nucleotides.
- Subsequent steps include array hybridization and streptavadin-Cy3 staining, which is quantitatively detected on scanning.
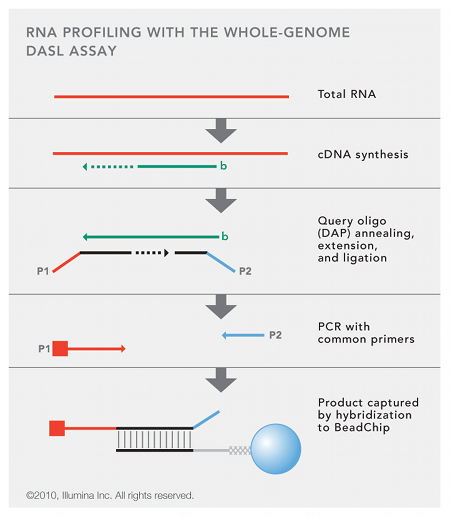
DASL Assay for degraded RNA (eg RNA extracted from FFPE samples)
- DASL = cDNA-mediated Annealing, Selection, extension and Ligation
- Probes span only 50 bases therefore suitable for degraded RNA
- Additional, unique probe sequences per gene increases the sensitivity of the assay
- Generation of expression profiles from as little as 100-200ng FFPE RNA
- The Whole-Genome(WG) DASL Assay Covers >24,000 transcripts.
- The assay combines unique PCR and labelling steps with gene-based hybridization and the whole-genome probe set of Illumina’s Direct Hybridization Assay.
- The Whole-Genome DASL Assay obtains high concordance with qPCR at a low per sample cost compared to other methods.
WG-DASL Method
- Convert total RNA to biotinylated cDNA
- Anneal cDNA to DASL assay pool (DAP) probe groups
- Upstream oligo with universal 5’ PCR primer sequence (P1) hybridises to targeted cDNA
- Extension of sequence and Ligation to downstream oligo with universal 3’ PCR primer sequence (P2)
- Resulting PCR template then amplified with universal PCR primers (P1 and P1)
- Product hybridised to BeadChip